Control and Prediction of the Organic Solid State
Results from the automated parallel crystallisation platform
Many systems have been studied using the automated parallel crystallisation polymorph screening platform in the University of Strathclyde.
Method
A full screen on a compound will typically involve approximately 600 distinct crystallisations from solution. This may require around 1kg of material and although smaller amounts can be used for more limited screens, the method is not applicable to amounts of less that 10g (for example a new compound where only a small amount has been prepared).
A solubility screen is carried out across a library of approximately 85 solvents with a diverse range of physicochemical properties. Based on the solubility screen, a full programme of crystallisations is planned; varying levels of supersaturation, solvent identity, agitation and method os inducing crystallisation. For each experiment, a measured amount of solid and solvent are added to the sample vial where they are heated and agitated to facilitate the dissolution process. Once solutions are saturated they are filtered, at elevated temperatures, into clean glassware prior to further heating by 1 – 2 deg C to remove any remaining particles that may seed the solution. Crystallisation may then be induced by controlled cooling, the addition of anti-solvent or by solvent evaporation via holding solutions under a constant vacuum. Finally, the recrystallised solid is recovered by filtration and analysed by powder X-ray diffraction. A fuller description of the experimental protocol is available in "An automated platform for parallel crystallization of small organic molecules", Florence, A.; Johnston, A.; Fernandes, P.; Shankland, N.; Shankland, K. J.Appl.Crystallogr. 2006, 39, 922-924.
This method has much higher reproducability of conditions than manual throughput methods, as it is so highly controlled. The exact conditions can be accessed through the instrument log files.
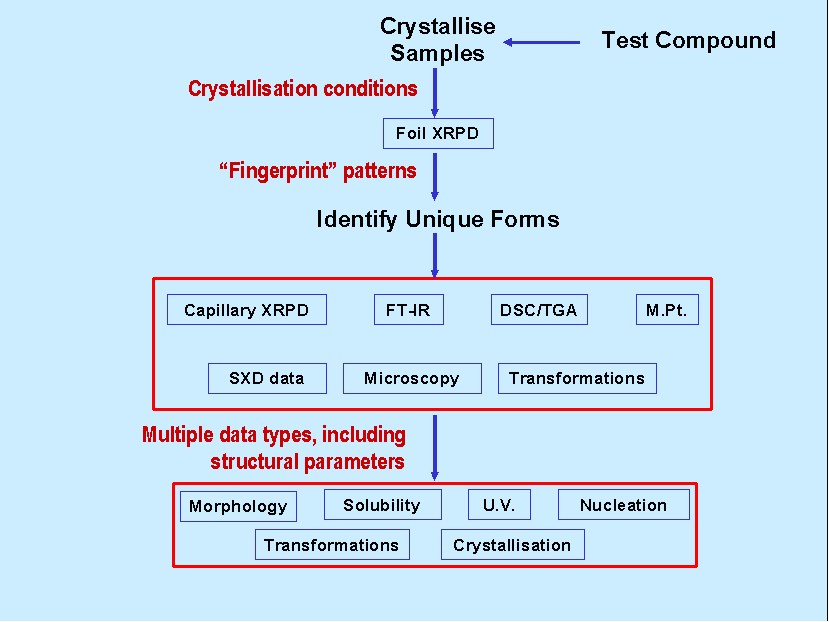
Figure 1: Schematic diagram illustrating a characteristic
workflow
Figure 1 illustrates a characteristic workflow for any particular compound of study. For each compound, full details of crystallisation conditions are entered into the database, the parameters include; compound, solid dose, solvent volume, solvent identity, saturation temperature, agitation and suitable descriptors for method of crystallisation (e.g. vacuum or cooling rate), see Figure 2.
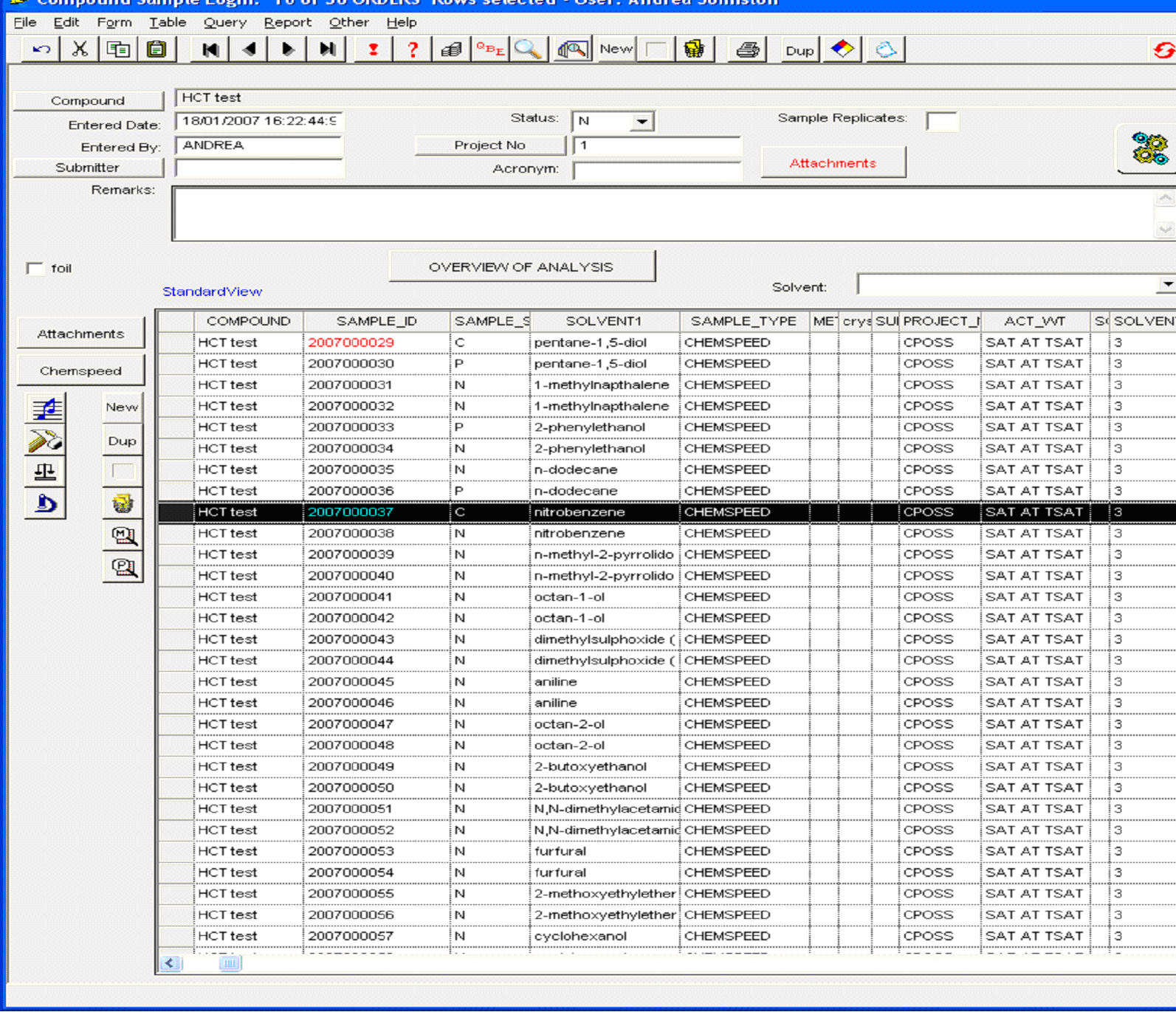
Figure 2: representative example of sample log-in window
containing crystallisation conditions, sample ID and sample status. (Click
on the image to enlarge and see the detail)
The system then generates discrete sample identification numbers which are used for all subsequent analyses (column 2) and in particular the details shown in Figure 2 are used to automatically generate a job file for XRPD analysis (Figure 3). Sample status (Figure 2, column 3) is another useful feature of the system which allows the user to recognise, at a glance, whether any one sample has been fully characterised as a known form and analyses are complete, or whether the sample requires further investigation and what type of experiments are required and have already been carried out.
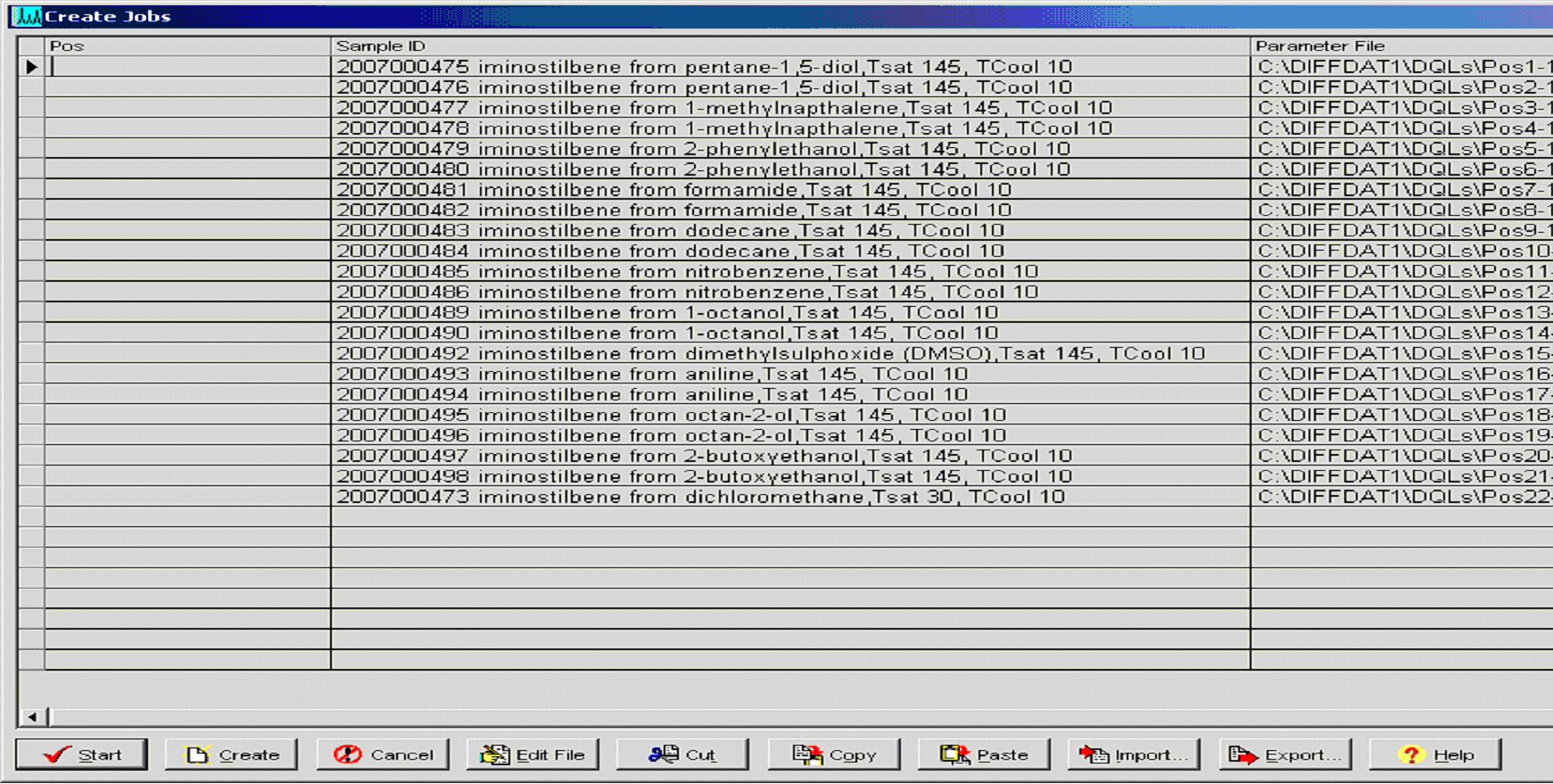
Figure 3: Job file for Bruker XRPD instrument automatically
generate using LIMS. (Click on the image to enlarge
and see the detail)
Associated data of each sample is stored as an attachment alongside data collection parameters coupled with related analysis. For example, a powder diffraction pattern will have associated data collection parameters including; instrument type (foil / capillary), 2? range, count times, step size, date etc., and analysis descriptions including; indexed unit cell parameters / figures of merit and whether the form is known or novel.
To summarise, our system provides automated secure storage for multiple data types and data sets generated for each compound and associated samples. The ability to search across the database on specific meta data that has been extracted from each data file facilitates more effective data handling and report generation.
Results
To date we have used the automated screening platform to investigate the following compounds. All forms observed in the screen are listed, along with publication details of new forms. Due to requirement for publications, a molecule may not be listed here until all output has been accepted by publishers. However, old systems are continually revisited, so further publications may ensue.
| Compound |
Polymorphs |
Solvates |
Co-crystal |
| Carbamazepine |
3 |
9 |
1 |
| Dihydrocarbamazepine |
3 |
6 |
0 |
| Hydrochlorothiazide |
2 |
7 |
1 |
| Chlorothiazide |
1 |
7 |
0 |
| R-1-phenylethylammonium-R-2-phenylbutarate |
2 |
0 |
0 |
| 3,4-dichloronitrobenzene |
0 |
2 |
0 |
| 3-azabicylcononane-2,4-dione |
1 (+ plastic phase) |
2 |
0 |
| Phthalimide |
0 |
0 |
0 |
Manual crystallisations
As an automated screen takes between two and ten weeks, the automated crystallisation team are supported by the small manual crystallisation team at UCL. The method is similar to that detailed above, but control and reproducibility within the laborotory is not as great as it is in Strathclyde. Powder diffraction is not routinely available at UCL as it is at Strathclyde, so IR is often used to identify samples and single crystal X-ray diffraction is used where appropriate.
Enter secure pages (For project members only - password required)
© UCL Chemistry Department 2004. This page was last updated on 17 August, 2017. If you have any problems with this page please email the WebMaster
